Data Scientist
MD Anderson Cancer Center
Ph.D. SUNY at Buffalo
Born September, 1988
S. Mostafa Sarayi
(Fardad)
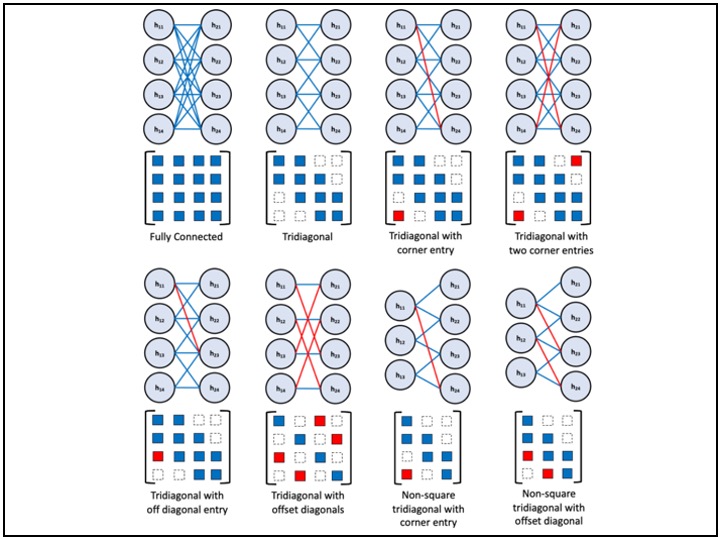
I am a Data Scientist at MD Anderson Cancer Center, formerly a Senior Statistical Analyst, with a background in Statistics, Data Science, Applied Mathematics (as a lecturer), Health Sciences, Bioengineering, and Mechanical Engineering. My passion lies in developing and deploying data-driven solutions for real-world problems, particularly in tech industry, healthcare analytics, computer vision, Nartural and Large Language modeling. I specialize in machine learning, statistical analysis, image processing, computer modeling, numerical simulation, and optimization.
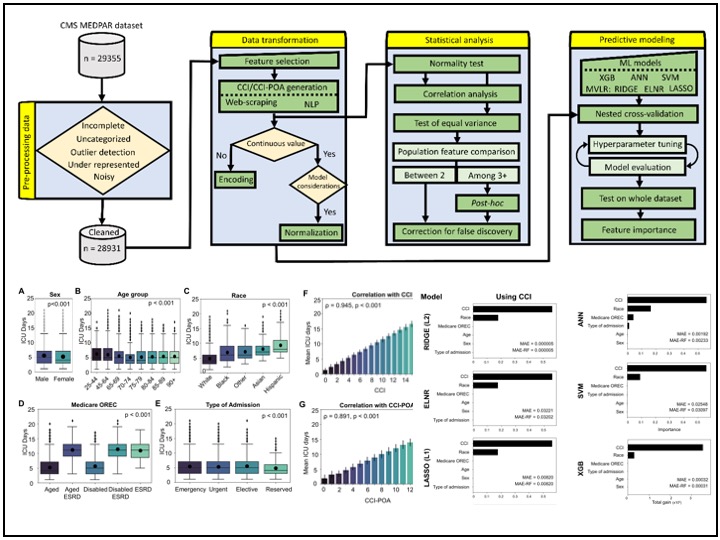
A key focus of my work involves handling complex medical data challenges, including highly imbalanced datasets in the cancer domain and across various modalities such as imaging, clinical records, and multi-source health data. I work extensively with cancer-related datasets, including national surveys, health claims data, cancer registries, clinical trials, and observational studies, ensuring that predictive models remain robust even in data-scarce or highly skewed scenarios.
At MD Anderson, I develop predictive models for cancer incidence, mortality, and risk assessment using machine learning and deep learning techniques. My work also involves generating microsimulation models to evaluate the cost-effectiveness and efficacy of cancer screening and treatment strategies. By integrating insights from diverse data sources, I contribute to evidence-based decision-making in oncology research and patient care.
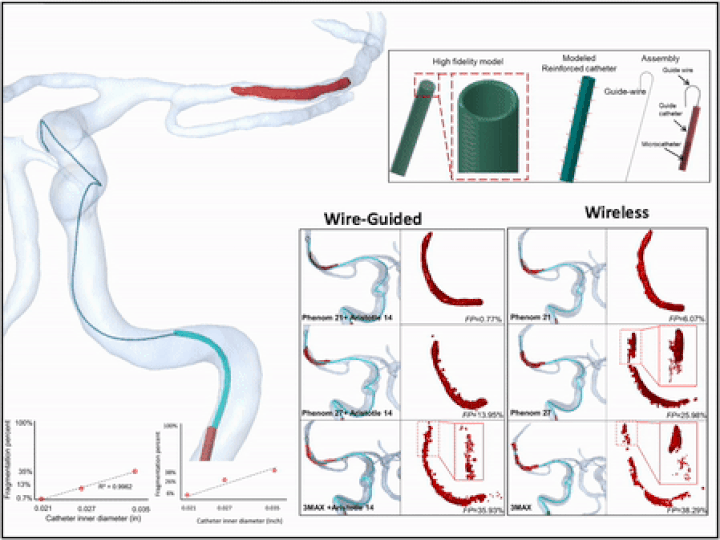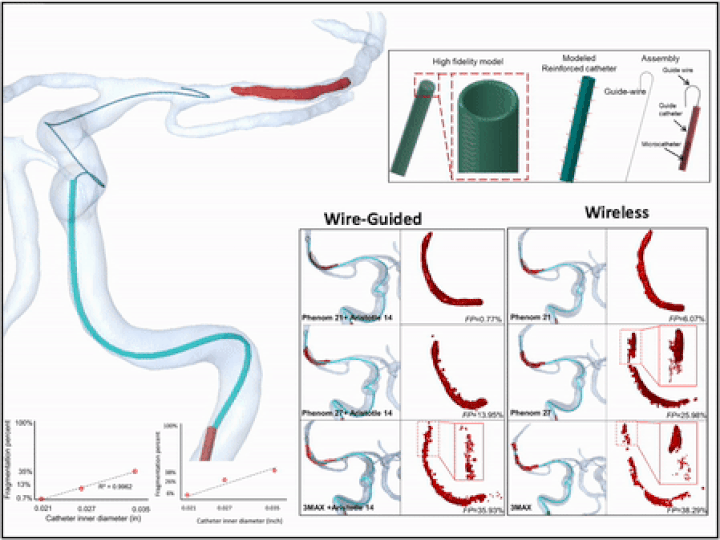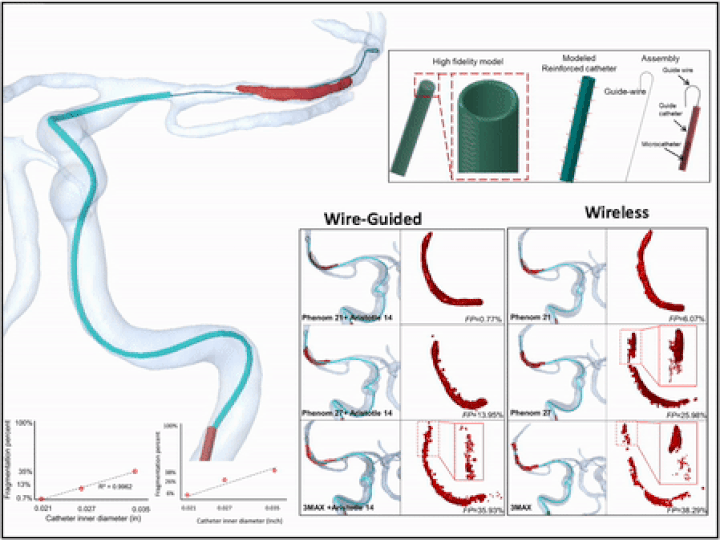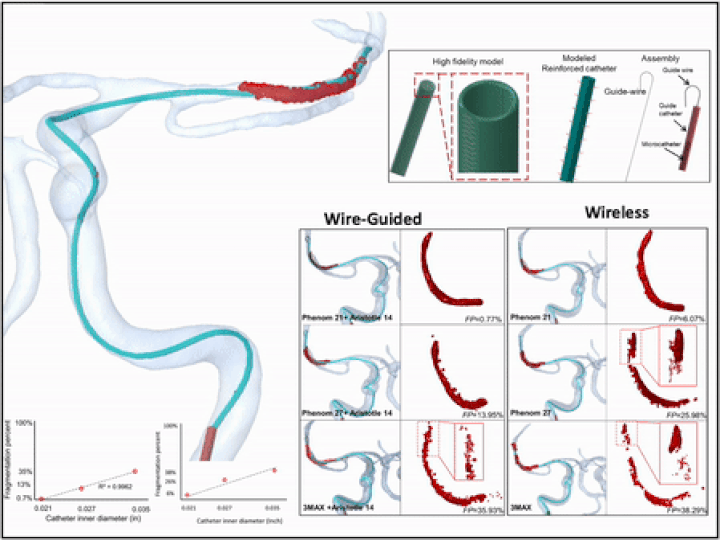
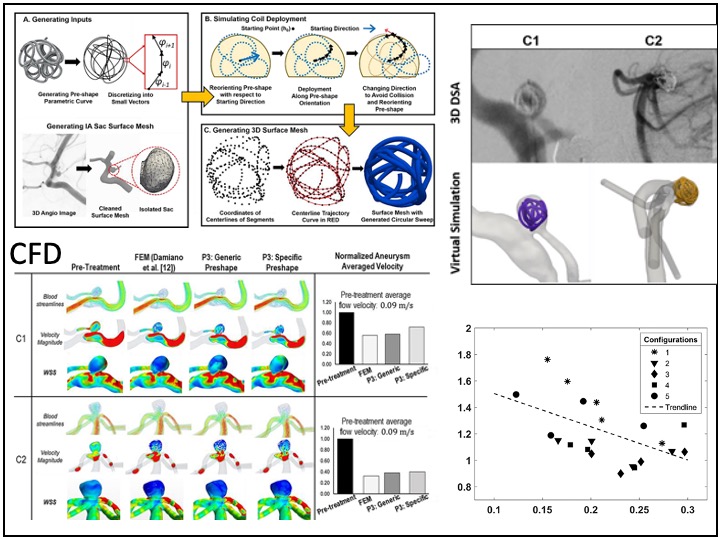
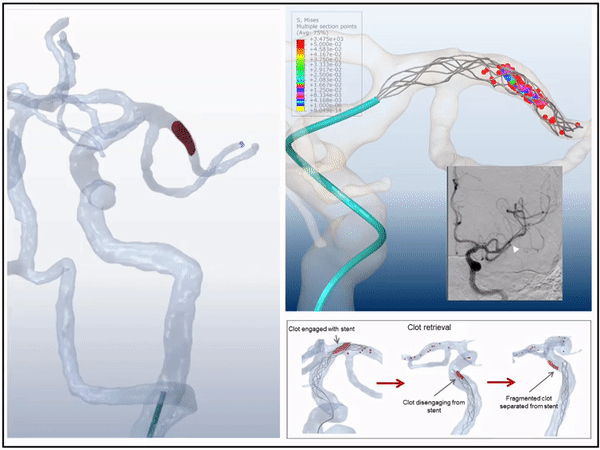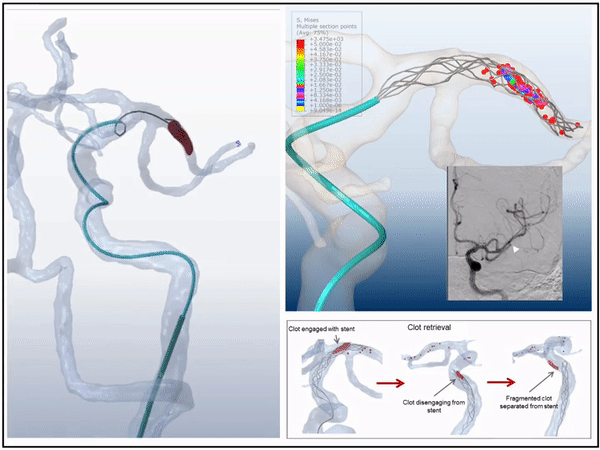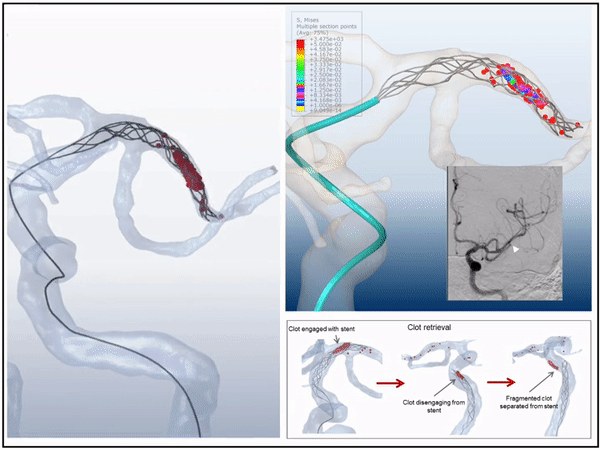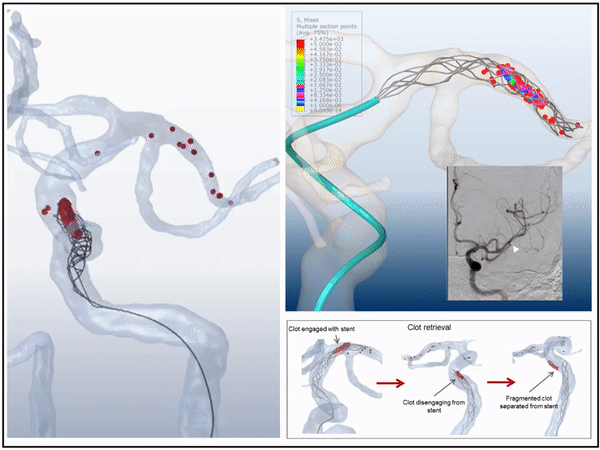
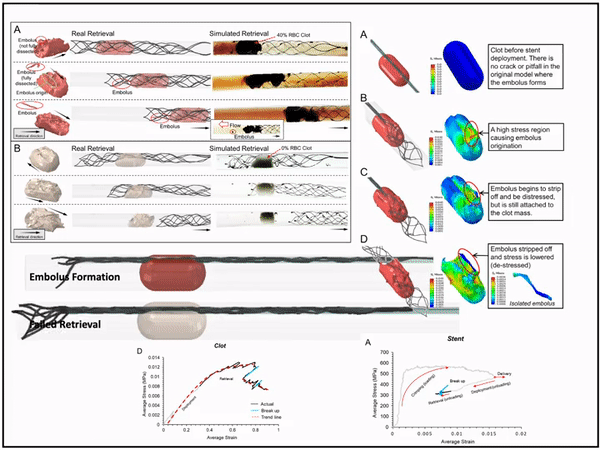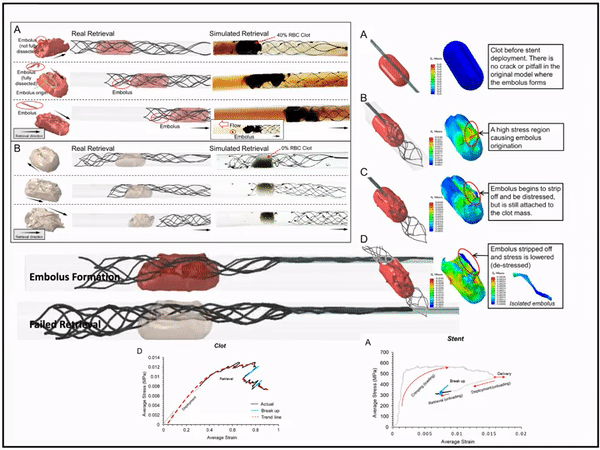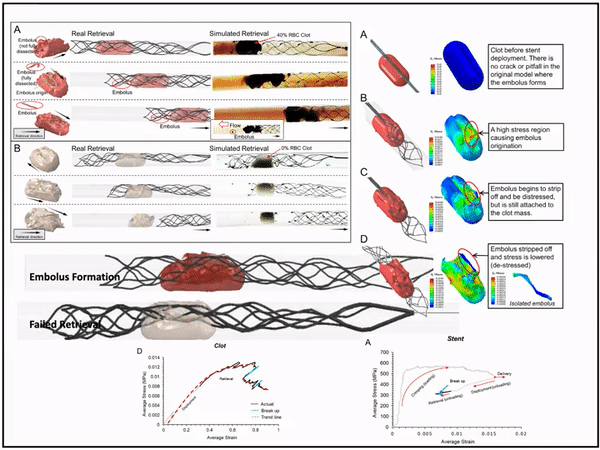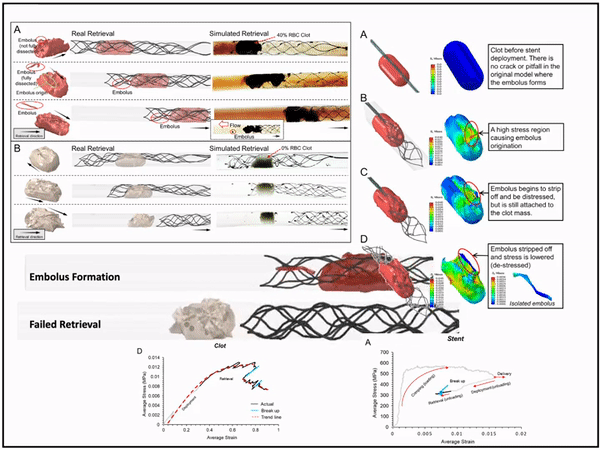
Previously, I served as a part-time technical consultant for Medtronic Co., where I led the development of numerical simulations and data-driven optimization models to enhance the efficacy of newly developed endovascular intervention devices.
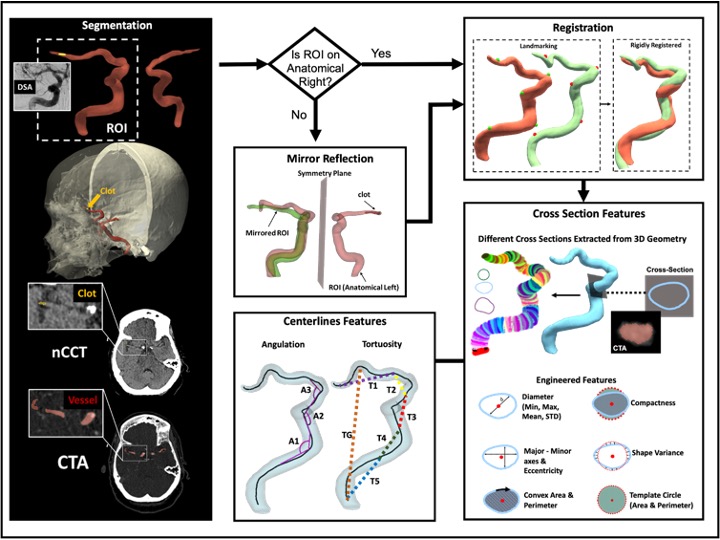
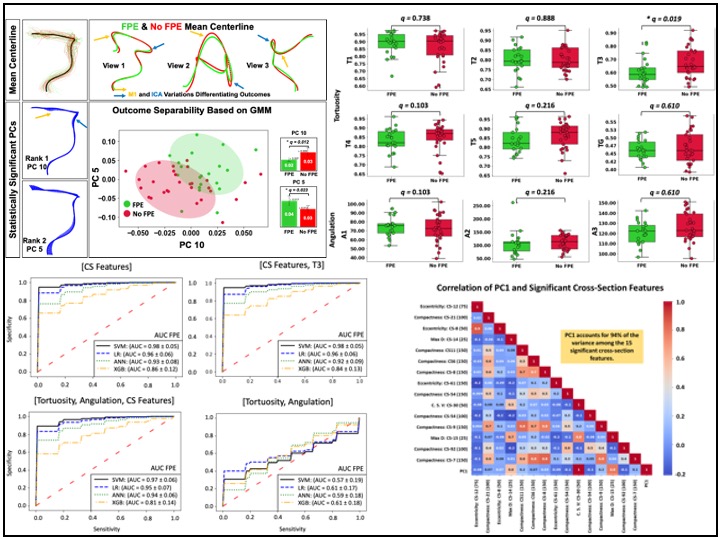
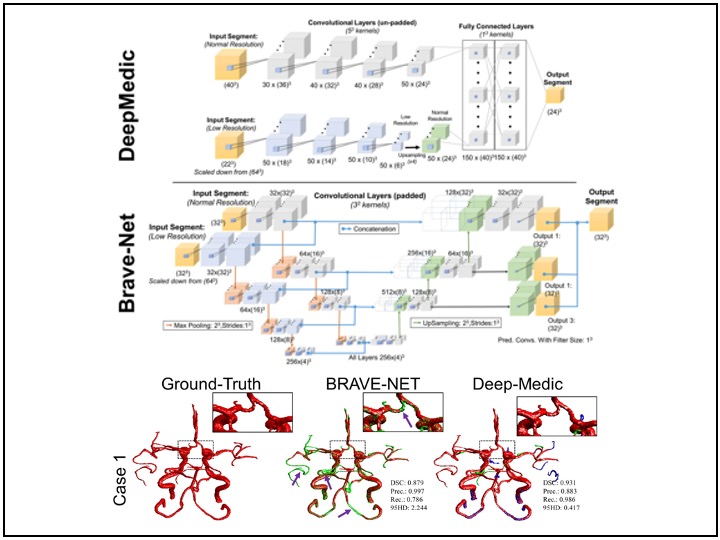
My experience in medical image analysis includes working with various imaging modalities such as digital images, CTA, nCCT, MRI, and Time-of-Flight MRA. I have developed automated segmentation, registration, feature extraction, statistical shape analysis, and machine learning models, contributing to improved diagnostic accuracy and patient outcomes. In addition to imaging, I have worked extensively with multi-institutional and nationwide patient datasets, including hospital records, insurance claims, clinical trials, and observational studies. My work in these areas integrates NLP, statistical modeling, feature engineering, and machine learning to extract meaningful insights that drive innovation in healthcare.
My research interests span healthcare AI, medical imaging, computational medicine, and big data in healthcare. I am particularly interested in improving model generalizability when working with imbalanced datasets, integrating multi-modal data sources, and advancing AI-driven solutions for oncology and medical decision-making.